- Home
- Applications
- Measure Cell Migration and Chemotaxis in Real Time
- Incucyte® Chemotaxis Assay
- Chemotaxis Protocols
- Detailed Adherent Chemotaxis Cell Migration Protocol
PROTOCOL
Detailed Adherent Chemotaxis Cell Migration Protocol
Detailed Demonstration Protocol
The following protocol is a detailed example designed to enable you to run a successful IncuCyte® Chemotaxis Cell Migration Assay. Note, that the protocol does not include a description of any experiments required for optimizing seeding density or surface coatings. Here we specifically describe the use of IncuCyte® ZOOM instrument for establishing and quantifying the inhibition of HT-1080 serum-induced chemotaxis in the presence of Cytochalasin D.
Download the detailed aherent protocol Request more information
HT-1080 Chemotaxis: Inhibition
Materials
- NucLight Red HT-1080 (Essen BioScience 4485) cells in culture (T75, 37°C, 5% CO2) in:
- F12 + 10% FBS + 1% Glutamax + 1% Pen/Strep. + 0.5 μg ml-1 Puromycin
- IncuCyte® ClearView 96-Well Cell Migration Plate (Essen BioScience 4582 or 4599)
- 2x IncuCyte® ClearView™ Cell Migration Reservoir Plate (Essen BioScience 4600 or 4601)
- 0.25 % Trypsin/EDTA (Life Technologies 25200)
- D-PBS (w/o Ca2+, Mg2+, Life Technologies 10010)
- Insulin-Transferrin-Selenium (Life Technologies 41400-045)
- Fetal Bovine Serum (Sigma-Aldrich F2442-500mL)
- Cytochalasin D (Sigma-Aldrich C8273)
Protocol
Day 0
- Remove serum-containing medium from culture and gently rinse twice with D-PBS.
NOTE: Culture should be below 80% confluence. A T-25 at 75-80% confluence generally yields enough cells to perform an entire 96-well Chemotaxis Cell Migration Assay. - Replace with serum-free F12 + 1x Insulin-Transferrin-Selenium (ITS). Return culture to incubator for 20 hours.
Day 1
- In advance, prepare a 3x stock of Cytochalasin D (e.g. highest cell treatment of 3 µM requires a stock concentration of 9 µM) and prepare 3-fold serial dilutions.
- Remove serum-free chemotaxis medium from culture and gently rinse with D-PBS.
- Harvest cells and perform a cell count (e.g., trypan blue staining + hemacytometer). Centrifuge the cell suspension (1000 RPM, 4 minutes) and resuspend the cell pellet in culture medium at 25,000 cells/mL. (Calculation: 25,000 cells/mL x 0.04 mL = 1000 cells per insert well).
NOTE: When performing a chemotaxis cell migration assay without treatment, the cell seeding volume is increased to 60 μL, thus the cell seeding stock is reduced to 16,666.67 cells/mL.
- Using a manual multi-channel pipette and reverse pipetting technique, seed cells (40 µL per well, 1,000 cells per well) into every well of the insert plate. Learn how to seed cells

- Add 20 µL of the prepared Cytochalasin D at 3x final assay concentration to the appropriate wells and mix by repeatedly pipetting up and down with a 30 µL volume.
- Allow the cells to settle at ambient temperature for 15 minutes then place the ClearView Cell Migration Plate at 37°C, 5% CO2 for 30 minutes in order to pre-incubate the HT-1080 cells in the presence of Cytochalasin D.
- Prepare F12 + ITS + 10% FBS as the chemoattractant, and F12 + ITS for the control wells.
- Using a manual multi-channel pipette, add 200 µL of the chemoattractant and control medium to the appropriate wells of the second reservoir plate.
- Carefully transfer the insert plate containing cells ± Cytochalasin D treatment into the reservoir plate containing medium ± chemoattractant. Learn how to transfer the insert

- Place the ClearView Cell Migration plate into the IncuCyte® ZOOM instrument and allow the plate to warm to 37°C for at least 15 minutes. After 15 minutes, wipe away any condensation that remains on the outside of the plate lid or bottom of the reservoir.
- In the IncuCyte® ZOOM software; schedule 24 hour repeat scanning (10x) for every 1-2 hours.
- a. Objective: Ensure 10x objective is installed.
- b. Vessel Type: Select ‘ClearView Cell Migration’
- c. Channel Selection: Select ‘Phase’ + ‘Red’ (800 ms acquisition time).
- d. Scan Mode: Select ‘Chemotaxis (Top/Bot)’ scan type and desired Scan Pattern
- e. Note IncuCyte® estimates a scan time of 20 min per plate (phase only) and 33 min per plate (phase and red); however, the actual scan time can take longer if condensation is not properly removed.
Example of Data Generated with the Detailed Adherent Chemotaxis Cell Migration Protocol
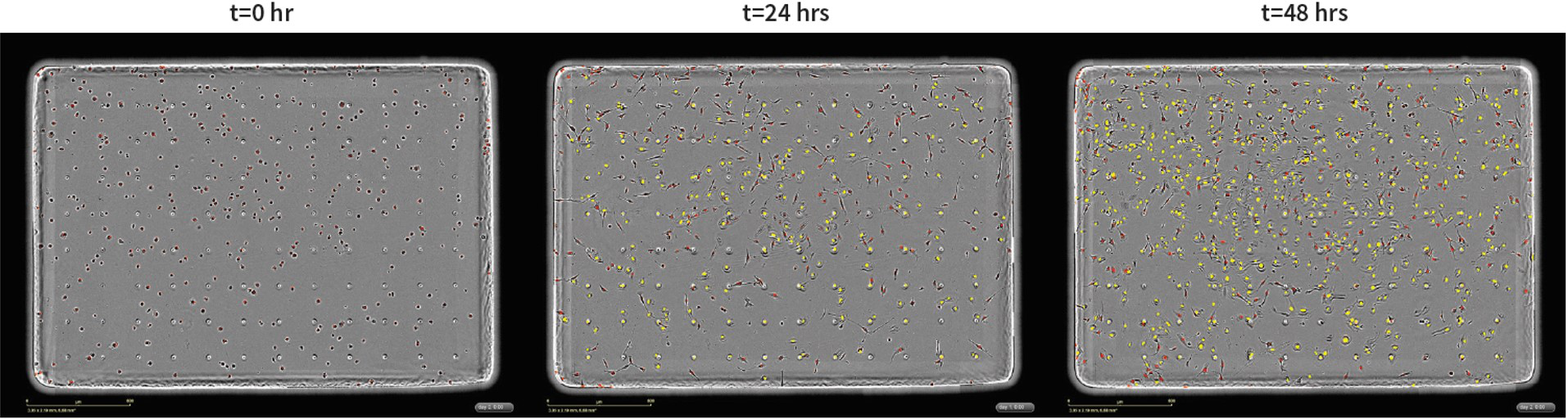
Representative images of NucLight Red HT-1080 cells migrating toward 10% FBS. Bottom-side fluorescence image segmentation mask (yellow) is blended with phase-contrast and fluorescence images acquired for NucLight Red HT-1080 cells migrating toward 10% FBS taken at t=0, 24, and 48 hours. Cells were seeded at 500 cells per well into an IncuCyte® ClearView 96-Well Cell Migration Plate.

Cytochalasin inhibition of NucLight Red chemotaxis. NucLight Red HT-1080 cells were pre-treated with varying concentrations of Cytochalasin D prior to the addition of 10% FBS chemoattractant into the ClearView reservoir plate. Images were analyzed for the cells that migrated to the bottom-side of the membrane and counted (n=4 per condition).
