- Home
- Applications
- Measure Cell Migration and Chemotaxis in Real Time
- Measure Chemotactic Migration and Invasion in Real Time
- Incucyte® Chemotactic Invasion

Incucyte® Applications
Chemotactic Invasion Assay for Live-Cell Analysis
What is directed cell invasion?
Chemotactic cell invasion is the directed movement of cells through an extracellular matrix in response to a chemical stimulus. It is a key component of cancer metastasis where invasive cells of the primary tumor degrade the underlying basement membrane, move toward the circulatory system and disseminate throughout the body. Metastatic potential is associated with tumor cells that have undergone epithelial-mesenchymal transition (EMT) and display a mesenchymal-like phenotype, which includes enhanced migratory capacity and invasiveness.
Cell-based assays enabling real-time visualization and detailed phenotypic analysis of chemotactic cell invasion are critical to further understand the processes involved in cancer metastasis and the discovery of treatments that block metastasis.

4th Edition! Live-Cell Analysis Handbook
A guide to real-time live-cell imaging & analysis

Application Note:
Gain additional insights into the mechanism of immune cell killing by combining live cell analysis and flow cytometry into a single workflow
Download Your CopyOverview
Introducing the Incucyte® Chemotaxis Invasion Assay
A fully integrated solution enabling real-time visualization and automated analysis of chemotactic cell invasion in a 96-well format – all inside your tissue culture incubator.
- Investigate treatment effects on directed tumor cell invasion
- Quantitatively assess metastatic potential and visualize morphological changes associated with epithelial-mesenchymal transition (EMT)
- Measure chemotactic cell invasion through 3D extracellular matrix gels of your choice.
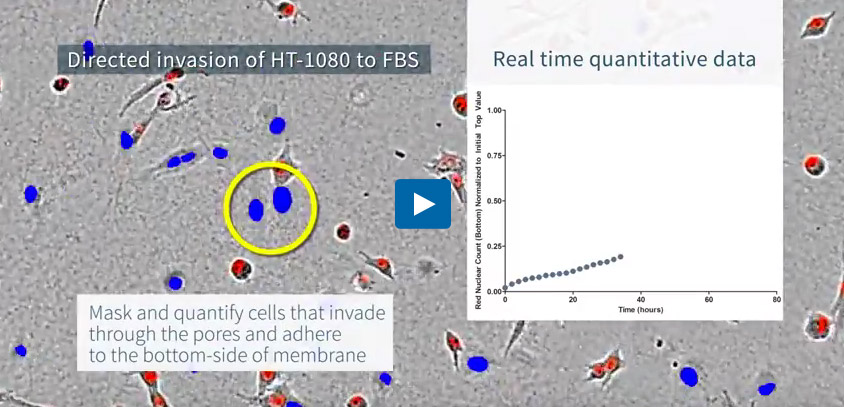
Directed invasion of HT-1080 fibrosarcoma cells through a 3D matrix of basement membrane extract (BME) toward serum. Visualized using an Incucyte® Clearview 96-well Plate. HT-1080 cells expressing a nuclear restricted red fluorescent protein (Incucyte® Nuclight Red Lentivirus) were visualized and quantified in real time as they invaded through BME gel using the Incucyte® Live-Cell Analysis System. Integrated Incucyte® Chemotaxis Analysis Software Module enables easy quantification of cells that have moved through the membrane pores and adhere to the lower surface (indicated as blue objects).
Chemotaxis Cell Invasion Quick Guide

Key Advantages
Key advantages of the Incucyte® Chemotaxis Assay
- Real-time visualization and fully automated analysis of tumor cell invasion
- Investigate treatment effects on tumor cell invasion
- Quantitatively assess metastatic potential
- Highly reproducible 96-well kinetic assays suitable for screening
Real-time visualization and fully automated analysis of tumor cell invasion
- Monitor every cell in your experiment, assess morphology and gain phenotypic insight from images and movies
- Automatically quantify cell invasion without the need for fixing, staining or cell scraping steps
Monitor chemotactic invasion in real time through your choice of 3D extracellular matrix gel. Time-lapse images of a nuclear red labeled HT-1080 tumor cell (black arrow) invading through a matrix of basement membrane extract toward a pore leading to serum (orange circle). The cancer cell extends into and penetrates, the matrix moving toward and through the pore. In the final image, the cancer cell has passed through the pore and has adhered to the underside of the membrane (blue circle) where it is quantified.
Investigate treatment effects on tumor cell invasion
- Characterize pathways involved in directed cell invasion through pharmacological profiling and genetic manipulation
- Investigate differential effects on cell migration and invasion in the same plate
- Visualize morphological changes associated with epithelial-mesenchymal transition (EMT)
Profile inhibitors of tumor cell invasion. Incucyte® high-definition images of nuclear red labeled HT-1080 fibrosarcoma cells migrating across a 2D substrate and invading through a 3D basement membrane extract (BME) biomatrix in response to a serum gradient – note the differential morphology and invasive filopodia-like projections that extend into the ECM. Time course plots reveal that cell invasion through the BME matrix is slower than migration and that invasion, but not migration, is inhibited by the matrix metalloproteinase inhibitor, GM6001 in a concentration dependent manner.
Quantitatively assess metastatic potential
Cross compare the invasive capacity of cell types in a single plate (e.g. isogenic pairs, tumor biopsies)
Quantify metastatic potential. Comparison of the migration and invasion time-course profiles for a highly invasive (HT-1080 fibrosarcoma) and weakly invasive (NIH-3T3 fibroblast) cell type in the presence of increasing 3D gel density (basement membrane extract). Note that the NIH-3T3 cells are effectively unable to invade through the ECM, but do migrate in the absence of the 3D biomatrix. NIH-3T3 cells show less metastatic potential than HT-1080 cells.
Highly reproducible 96-well kinetic assays suitable for screening
- Measure chemotactic cell invasion in up to six 96-well plates at once (576 wells of data at every time point)
- Set up and walk away – fully automated image based quantification
- Compatible with standard automation and liquid dispensing devices
Robust 96-well assays. Representative 96-well microplate graphs showing HT-1080 invasion through a 3D basement membrane extract biomatrix towards serial dilutions of serum to illustrate inter-plate reproducibility. Z’ values ranged from 0.7 to 0.8 for four replicate plates over three days. Corresponding concentration-dependent response curves to FBS provided reproducible measurements of the pro-invasive effects of serum (EC50 value range 1.5% to 3.4%) within and between days.
FAQs
Chemotaxis FAQs
Membrane-based transwell systems for chemotaxis assays are capable of maintaining chemoattractant gradients for a few hours, but beyond that they collapse, leading to equimolar amounts of chemoattractant on both sides of the membrane. ClearView 96-Well Chemotaxis Plates rely on 96 tiny pores (8 µm in diameter) to set up and maintain the chemoattractant gradient for 72 hours. Since chemotaxis migration assays rely on chemical gradients to differentially induce migratory behavior in cells, maintaining the chemoattractant gradient is essential.
Yes, rather than just citing the reduction of cells on the surface of the membrane, the IncuCyte™ ZOOM can provide real-time analysis of the movement of cells through the pores to the underside of the well. It simultaneously provides live-cell analysis of both surfaces of the insert, enabling morphological analysis of the cells before, during, and after migrating through the pores.
Chemotaxis assays analyze cell migration in response to a chemoattractant, a process that guides cell trafficking in vivo. Examples of chemotaxis assay applications include the analysis of chemotactic migration across substrate surfaces, chemotactic invasion through 3-D biomatrix gels, and chemotactic transendothelial migration of leukocytes (extravasation or diapedesis). Studying chemotaxis requires the generation of chemotactic gradients, as cells travel from areas with one concentration to areas of another (high concentration to low concentration, or low concentration to high concentration). Some chemotaxis assays rely on membranes to maintain these chemoattractant gradients, but those gradients are unstable and they rapidly collapse. ClearView 96-Well Chemotaxis Plates enable gradient maintenance for at least 72 hours -- Something other chemotaxis assay systems can’t claim.
Learn more about the ClearView method for chemotaxis assays
Leukocyte extravasation is the process by which leukocytes in the circulation stop rolling along vascular endothelium and migrate across the endothelium in response to inflammation. In order for any system to accurately recapitulate transendothelial migration, it must first involve the generation of a monolayer of endothelial cells over top of the pore-based insert. Once confluence of the endothelial cells is established, the leukocytes can be carefully added to the culture, where they will extravasate between the endothelial cells in response to the chemoattractant in the reservoir.
View the IncuCyte™ transendothelial migration assay protocols
The most common effector cells used in the study of trans endothelial migration of leukocytes in vitro include primary T cells, neutrophils, and Jurkat cells. The most commonly used cells to recapitulation the endothelial cell layer are HUVECs. Other cells not listed here that exhibit the ability to extravasate through the endothelium may work with this system, but the protocol – including the choice of endothelial monolayer – will need to be experimentally determined.
While immune cells are non-adherent, their interaction with ECM on the substrate is essential for their migration. Immune cells do not migrate on uncoated surfaces, but can readily migrate in the presence of appropriate ECM proteins. Examples of substrate coatings that are compatible with immune cell chemotaxis include ICAM-1, fibronectin, and Matrigel®/FBS.
Ordering Info
Incucyte® Chemotaxis Analysis Software Module
An add-on software module for the Incucyte® Live-Cell Analysis System. Analyze label-free and fluorescently-labeled chemotactic cell migration images acquired using the Incucyte® Clearview Plate for chemotaxis. This software is required for analyzing chemotaxis using the Incucyte® Live-Cell Analysis System.
Incucyte® Clearview 96-Well Plate for Chemotaxis
The Incucyte® Clearview 96-Well Plate provides an optically clear surface for label-free imaging and analysis of chemotactic cell migration within the Incucyte® Live-Cell Analysis System. The plates are required for analyzing chemotaxis using the Incucyte® Live-Cell Analysis System.